Ultimate Awesome of Awesomeness Mass Spec. Calc. + Builder
Installing the Mass Spec. Calc. App
The Ultimate Awesome of Awesome Mass Spec. Calc. is available on either Mac or Windows. To install, download the zip file corresponding to your system and unzip. On PC, double click on massSpecMain.exe to run the program. On Mac, double click the Mass_Spec_App to run.
Developed by Ryan K. Spencer, email rspencer@uci.edu regarding questions/comments/bugs.
Building a Sequence
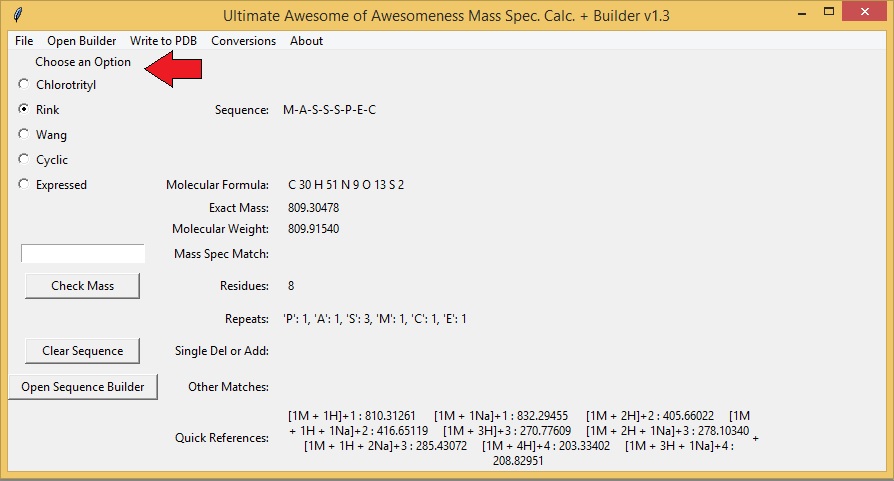
After running the App you should see the following screen (see below). There are a couple ways to Build a Sequence in the Mass Spec App : using the "Sequence Builder" window or loading the sequence from a text file (see section Save/Load Sequence). To build using the "Sequence Builder" window either click on the "Sequence Builder" button (bottom left of the main window) or select "Sequence Builder" from the "Open Builder" tab at the top of the app.

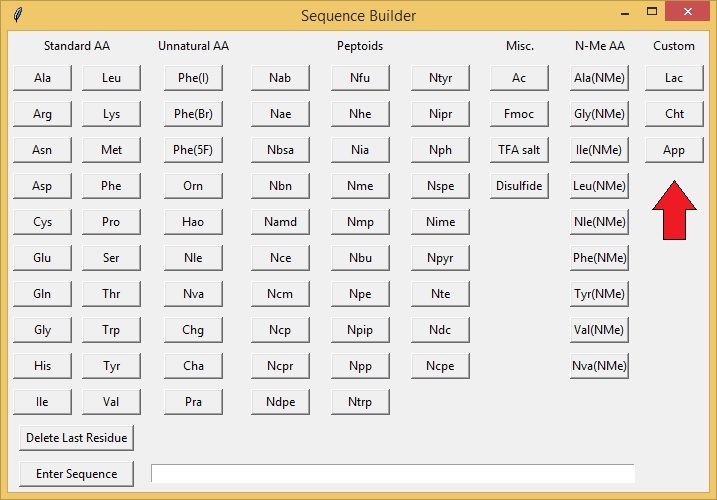
This will open a second window containing all the amino acid and peptoid monomers that are available for building a sequence. You may click on any button to add a residue to a sequence. Sequences are built from N to C termini. You may also add "Custom" buttons for monomers that are not shown in the window (See section Add/Delete Custom Buttons). You can also enter your sequence as a string (ex. MASS SPEC) in the "Enter Sequence" section at the bottom of the window. To delete a single residue from the end of the sequence hit the "Delete Last Residue" button.

The sequence is automatically updated on the Main Window as buttons are pressed or as the sequence is entered as a string. The main window will display the Molecular Formula, Exact Mass, Molecular Weight, Number of Residues, and Repeats. Choose how the polypeptide was made (synthetic or expressed) to display the correct mass (default displays the C-terminal end as an acid). You may clear the entire sequence by clicking on the "Clear Sequence" button.
Save/Load a Sequence.
To Save or Load a sequence, choose File -> Save Sequence or Load Sequence. The sequence is saved as a simple text file that can be loaded at a later time. You may also load a text file containing your sequence. For natural amino acids you may use single letters (ex. A) or triple letters (ex. Ala). For unnatural amino acids you can use the three or more letter code corresponding to a particular residue (ex. Nce or Phe(I)).
Comparing Mass Spec Data to Sequence
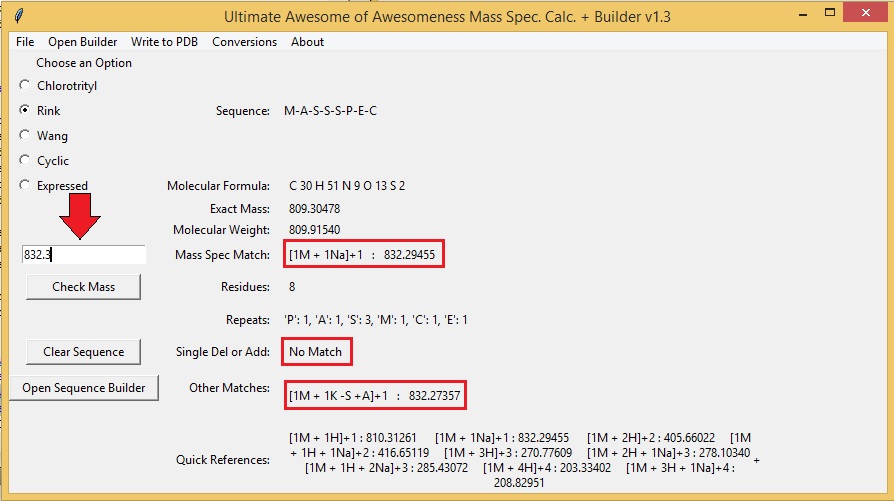
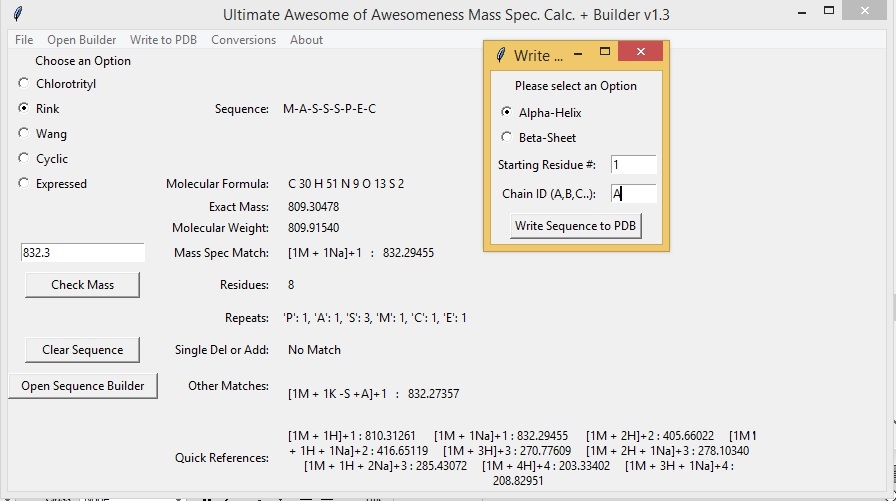
To compare mass spec. data to your sequence simply type the value of the observed peak into the text box above the "Check Mass" button and press the "Check Mass" button. The program will compare the observed mass spec peak to the theoretical mass spec peaks of multiple mass spec adducts. The program is capable of calculating H, Na, and K adducts with multiple charges and combinations as well as oligomers (dimers, trimers, tetramers, etc.). The results are displayed next to "Mass Spec Match". The program also calculates and compares the mass spec you enter to possible addition or deletion adducts, as well as double addition/deltions and single deletion and addition adducts. A quick references of the theoretical peaks for your sequence are displayed at the bottom of the window.
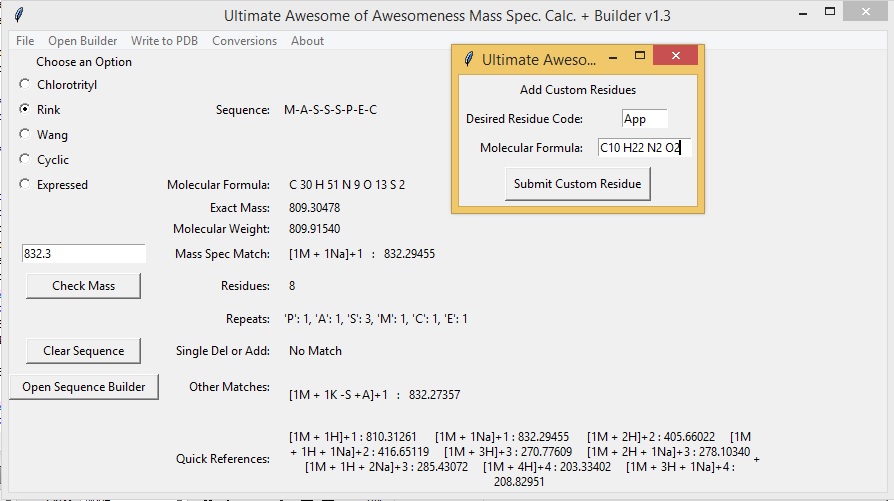
Add/Delete Custom Buttons
The Mass Spec App comes with over 80 residues/fragments to choose from but there may be residues/fragments that you use that are not listed. To add a custom residue/fragment to the Sequence Builder choose Open Browser - > Add Custom Button. This will generate a second window that will allow you to add custom residues. Type in a desired residue code and the molecular formula of the residue as an acid and free amine. Entries must contain an element and a number (ex. C 1 O 2). Click "Submit Custom Residue" to save the residue. Newly added buttons will appear on the far right side of the Sequence Builder Menu.
Generate a PDB file from a Sequence.
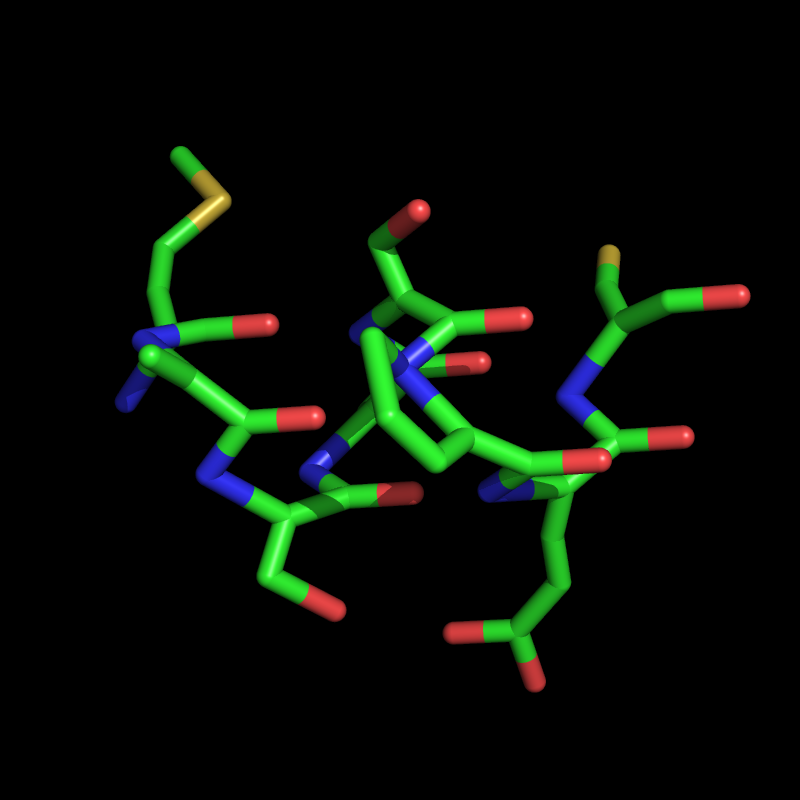
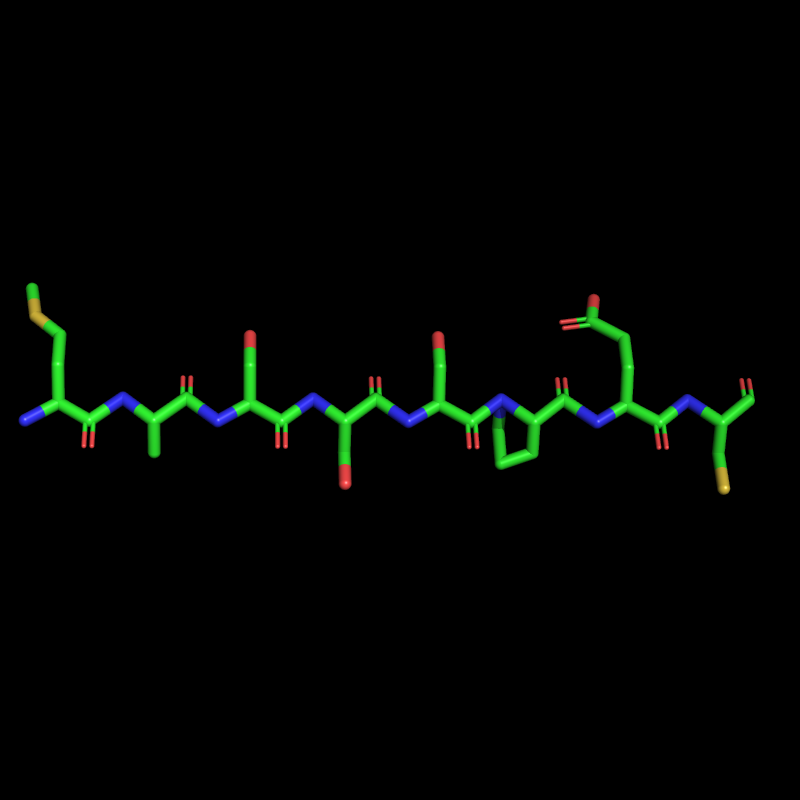
The Mass Spec App is also capable of generating a "crude" 3D structure based on your sequence. *** *NOTE* The PDB coordinates generated do not reflect ideal angles or bond lenghts, further minimization should be performed in such programs as Pymol or Maestro to obtain more accurate structures.*** You have two basic choices: an alpha-helix or a beta-sheet. Currently all natural amino acids, N-Me amino acids, and unnatural amino acids can be used to generate a PDB structure. Only a few peptoid fragments can currently can be used and may only be used to generate a beta-sheet structure. "Hao" is also limited to a beta-sheet conformation. To generate a PDB select "Write to PDB" -> "Write PDB file". You may select alpha-helix or beta-sheet and enter a starting residue number and a chain letter (default values are 1 and A). Push the "Write Sequence to PDB" to generate the PDB file.
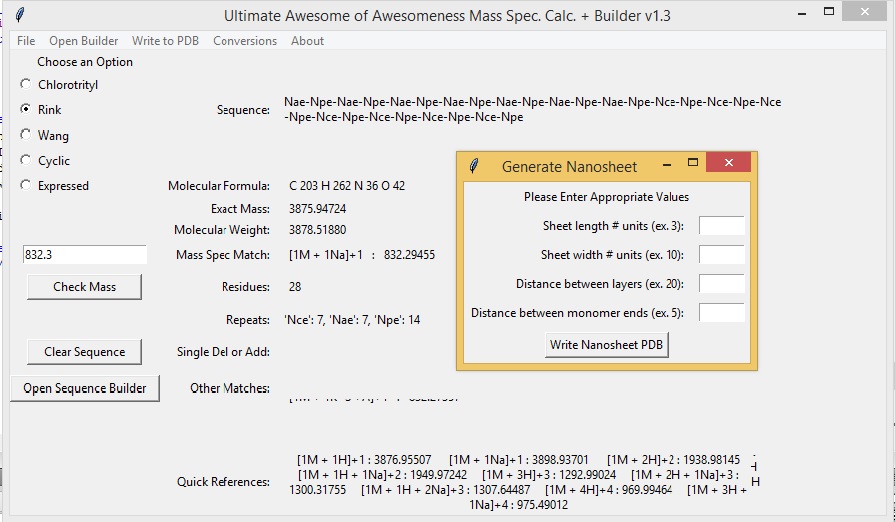
Generate a Nanosheet PDB file from a Sequence.
The Mass Spec App is also capable of generating a 3D nanosheet based on your sequence. All natural and unnatural amino acids and a few peptoids can be written to a PDB file. To generate a nanosheet select "Write to PDB" -> "What the Sheet?". An additional window will prompt you to enter length, width, distance between layers, and distance between monomer units.
<

<

<
