Current Areas of Research:
These are general descriptions, see “People/Projects” to learn more



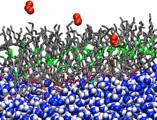
Atomistic modeling of interfaces relevant to atmospheric chemistry
The surfaces of aerosol particles are the site of unique chemistry that does not occur in the gas phase or bulk solution environments. For the purpose of quantitative modeling of chemical dynamics in the atmosphere and the contributions of aerosols to the Earth’s radiative balance, and hence, climate, it is essential to know the mechanisms and kinetics of reactions occurring at aerosol-air interfaces. Progress along these lines is limited by the difficulty of probing interfaces experimentally with molecular resolution. We are using force field-based and ab initio MD simulations to provide an atomistic view of molecular structure, electronic structure, and dynamics on the surfaces of model systems meant to mimic atmospheric aqueous and organic aerosol particles. In addition to providing new fundamental insight into the composition and physicochemical properties of aerosol surfaces and their interactions with water and atmospheric trace gases, our simulations are proving useful in the development of novel mechanisms for reactions that could be important in the chemistry of the atmosphere, including acid-base, redox, and photochemical reactions. Our work in this area is carried out under the auspices of AirUCI, an NSF-funded Organized Research Unit, headed by UCI Professor Barbara Finlayson-Pitts, which includes several other UCI faculty, partners at three national labs (Pacific Northwest, Lawrence Berkeley, and Lawrence Livermore), and international associates from New Zealand and the Czech Republic.
Structure and dynamics of proteins in membranes
Membrane proteins comprise roughly a quarter of the genome and the majority of pharmaceutical targets, but relatively few structures of membrane proteins are known, and structure prediction using methods developed for soluble proteins is not yet effective. In order to predict their structures with acceptable accuracy, it is necessary to know how membrane proteins interact with and are assembled in fluid lipid membranes sandwiched between aqueous compartments. We have been developing methods for using MD simulations to refine low-resolution structures of lipid bilayers and membrane proteins using x-ray and neutron diffraction data. In addition, we have been running simulations of large systems (several hundred thousand atoms) containing membrane proteins of known structure in explicit lipid bilayers in order to develop a microscopic description of how the membrane distorts and conforms to embedded proteins, to catalog important protein-lipid interactions, and to identify key protein-lipid-water interactions that are relevant to membrane protein function. Our membrane protein research is carried out in close collaboration with experimentalists in the TEMPO (Theory and Experiments in Membrane Protein Organization) group at UCI, as well as other collaborators elsewhere in the U. S. and in Europe.
Mechanism of voltage-sensing in voltage-gated ion channels
Electrical signals in excitable cells (e.g., neurons) are generated by the flow of ions through protein channels in membranes. In the case of voltage-gated ion channels, ion permeation is controlled by the opening and closing of the ion conducting pores in response to changes in the transmembrane electrical potential. Kv channels are built as tetramers, with each monomer contributing a pore domain and a voltage sensing domain (VSD). The VSD contains the S4 helix, which is rich in positively charged residues, predominantly arginines, for high sensitivity to small changes in transmembrane potential. In spite of the availability of crystal structures and a wide variety of spectroscopic and functional data, the molecular mechanism of voltage gating is still not well understood. The details that remain to be worked out, which we are addressing using MD simulations, include the lipid exposure of the charged residues in the S4 helix, the establishment of the location of the VSD in the closed state of the channel, where and how much the VSD moves through the membrane during membrane depolarization, and the nature of the electromechanical coupling through which the motion of the VSD opens and closes the pore. We are also generating atomistic models for proton channels (Hv1) based on their homology with the VSDs of potassium channels, and using force field-based and quantum mechanical/molecular mechanical (QM/MM) MD simulations to elucidate the mechanism of proton conduction.
Coupling of the dynamics of proteins and their surroundings
Protein function requires motion over a wide range of time and length scales. Both temperature and the solvent environment affect the spectrum of dynamical fluctuations in a protein. We have been using MD simulations to study the influence of the environment (aqueous solutions, glass forming co-solvents, and membranes) on protein dynamics. One of the primary objectives of this work has been to elucidate the effects of the environment on the protein dynamical transition from an inactive, glass-like state at low temperature to the functional, liquid-like state at high temperature. This transition, which is ubiquitous in biopolymers, has been of great interest for more than two decades in large part because of its connection to protein function, which for many proteins ceases when they go into the glass-like state. It has been recognized for some time that the solvent environment plays a key role in determining the transition temperature, and the amplitudes of motion in the liquid-like state, but the details of the protein-solvent coupling are still being worked out. In addition to its intrinsic value, this information is key to understanding how certain organisms protect themselves against low temperature and dehydration, and has applications to biopreservation in the food and pharmaceutical industries. The lipid composition-dependence of membrane protein dynamics is relevant to a wide variety of biological processes that rely on transmembrane signalling.