In this exercise we will learn to visualize and build molecules using PyMOL. First, we will visualize structures of caffeine and DNA. Then we will build structures of ibuprofen and cocaine.
1. Molecular Reprentations.
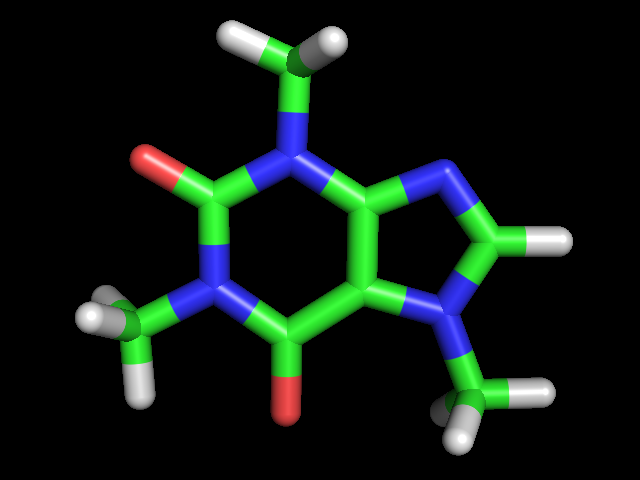
a. Download the structure of caffeine clicking on the linked caffeine.pdb file and open it using PyMOL.
b. Use the Show (S) button on the right hand side of the screen to show caffeine as sticks.
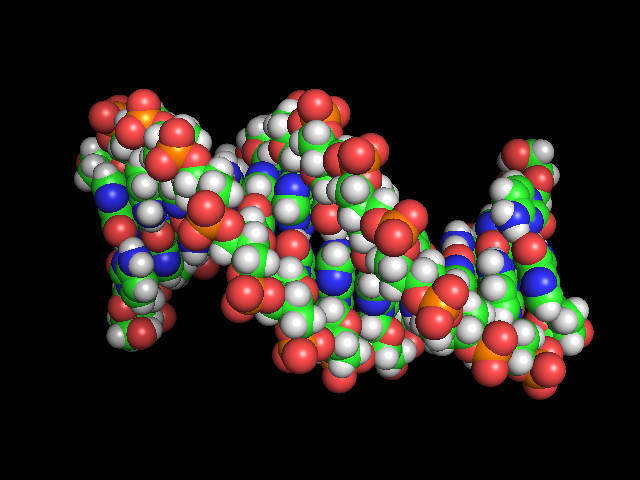
c. Download the structure of a molecule of DNA called the Drew-Dickerson dodecamer by typing fetch 2DAU in the command line. This will get a structure of a DNA molecule from a databank called the RCSB.
d. Display the structure of DNA as spheres using the the Show (S) button on the right hand side of the screen.
2. Building Molecular Models
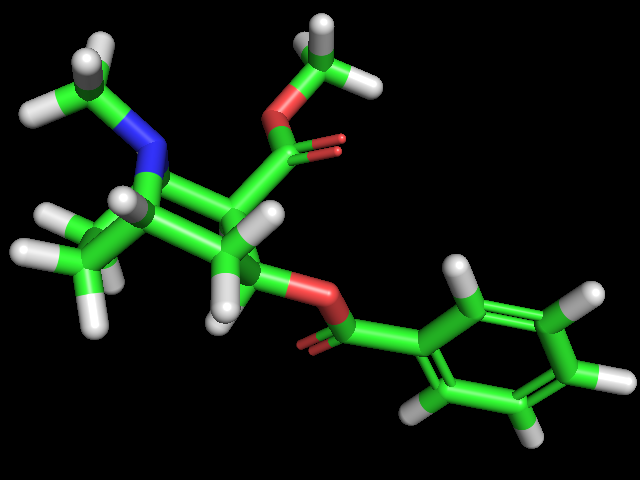
a. Build a molecular model of the (R)-enantiomer of ibuprofen. Use Builder menu and templates for benzene rings, CH4 groups, and C=OO groups to make the molecule. When you are done, use the Clean function to make the bond lengths and angles correct and to allow the molecule to adopt a reasonable conformation.
b. Build a molecular model of cocaine. Use the Builder menu and templates to make the cocaine molecule. Use the templates, Bonds, and Fix H features as needed during the building process. Use the clean function as needed along the way and use it at the end to make the bond lengths and angles correct and to allow the molecule to adopt a reasonable conformation.