Design of Ligands that Rescue Function in Mutant Forms of the Tumor Suppressor p53: We are part of a large, multidisciplinary collaboration that is studying how to "rescue" activity in mutant forms of the tumor suppressor p53. Our role is to design and synthesize small molecule ligands that bind to p53 and stabilize the core domain of some of the most common mutants found to be associated with human cancers. We employ a combination of de novo ligand design and library synthesis to search for ligands that ultimately might be useful in chemotherapy by restoring the function of mutant p53.
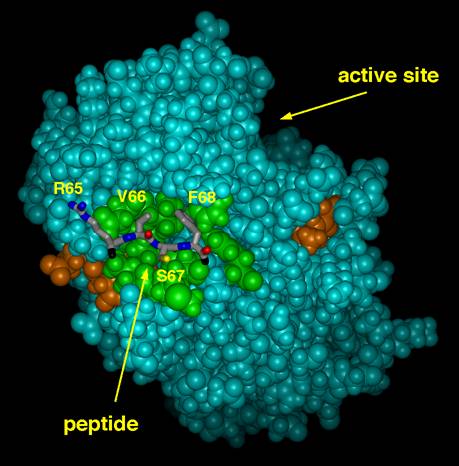
Phosphatase Activity Modulators: Substrates bind to the active site of protein phosphatase 1 (PP1) with relatively little sequence selectivity. In direct contrast, a wide variety of regulatory proteins interact with PP1at an allosteric regulatory site that recognizes a "loose" consensus sequence of R-V-X-F in the binding protein. In addition to localizing the phosphatase to specific cellular locations, this binding interaction serves to tailor phosphatase activity to each locale, increasing activity towards the relevant substrates and decreasing it towards others. This behavior is recapitulated by small RVXF-containing peptides, suggesting that it may be possible to design small molecule competitive ligands for the regulatory site, i.e., surrogates for the regulatory proteins that would "recruit" active PP1 into the cytosol and thereby lower the overall phosphorylation state of the cell. This possibility constitutes a completely new way of controlling signaling pathways that would be functionally complementary to the much more extensively explored stratagem of inhibiting kinases.