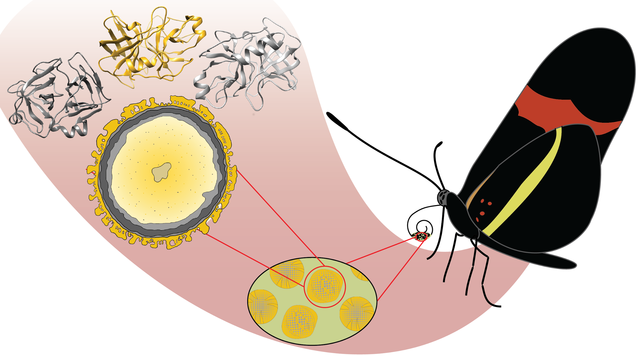
The Martin lab has published a paper in collaboration with the Butts and Briscoe groups describing the evolutionary history and function of the serine protease cocoonase in Heliconius butterflies. Moths use this protease to eclose from their silk cocoons. Butterflies do not make cocoons; they instead emerge from a chrysalis. Therefore, the presence of multiple isoforms of the cocoonase was a biological mystery. The Briscoe lab determined that these enzymes are involved in pollen feeding, which is particular to Heliconius butterflies. It was initially hypothesized that the different isoforms were used for digesting different protein substrates, however molecular modeling and structural analysis indicated that the active sites and specificity pockets of all the cocoonase isoforms are similar to trypsin. Instead, the cocoonase isoforms differ primarily in their surface residues, reflecting their need to enter different chemical environments within the complex biological milieu of pollen grains.
Read their paper here.
